Collaborative Research Centre 1648
Emerging viral infections pose a major threat to global health, as exemplified by the recent outbreaks of Ebola, MERS and Mpox, as well as the COVID-19 pandemic. These threats arise from the large number of potentially pathogenic viruses found in animal reservoirs and the associated potential for these viruses to jump to humans in spillover events. The initial lack of human-specific immunity to these viruses, the lack of antiviral treatment options, insufficient knowledge of replication, tropism, and host and viral structural targets, and the potential for rapid spread make them a threat. In preparation for future outbreaks, WHO has defined a list of priority diseases associated with specific viral infections, based on their epidemic potential and the lack of preventive and therapeutic countermeasures.
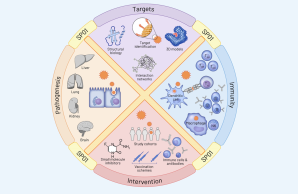
The Collaborative Research Center 1648 will address knowledge gaps in four critical research areas for scientific preparedness against emerging viral infections: Pathogenesis (A), Target Characterization (B), Immunity (C), and Intervention (D). Advanced cellular, structural, computational and chemical biology techniques will be used to identify critical virus-host interactions and to understand the molecular mechanisms underlying infection processes. The consortium will dissect vital aspects of both immune defense and immune pathology, addressing open research questions in vaccine design that can be translated to other emerging viruses and inform research and clinical strategy for intervention against future outbreaks.
This research consortium brings together a multidisciplinary team of 26 scientists from basic and clinical research to address the major challenge posed by emerging viruses through 16 research projects, one service project, and one outreach project.All PhD and MD students are automatically enrolled in the Integrated Research Training Group (iRTG). The goal of the iRTG is to train future leaders in the global fight against potential pandemics.
A particular strength of the collaborative initiative is how it synergizes and integrates the full spectrum of research approaches in specific emerging viral infections, structural biology and biophysical characterization of biomolecules and their interactions, immune defense mechanisms underlying inflammation and immunopathology, and early drug discovery and vaccine research. Molecular resources, workflows and innovative animal models will be established, resulting in a comprehensive technology toolkit for rapid application to a future "Disease X" (defined by the WHO as caused by an unknown pathogen with epidemic potential). By addressing one of the most critical research challenges of our time, this novel pipeline approach will ensure a rapid and strategic response to an emerging viral threat.
Press release
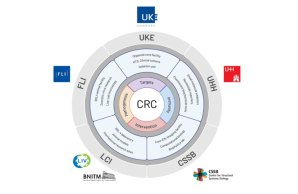
Participating Institutions
University Medical Center Hamburg-Eppendorf (UKE)
Bernhard Nocht Institute for Tropical Medicine
Centre for Structural Systems Biology
Integrated Research Training Group “Understanding emerging and re-emerging viruses”
All PhD and MD students are automatically enrolled in the Integrated Research Training Group (iRTG) "Understanding Emerging and Re-emerging Viruses".
The iRTG is closely aligned with the focus of the CRC "Emerging Viruses" and trains M.D. and Ph.D. students as future experts in emerging viral diseases. This multidisciplinary program, in collaboration with other graduate programs on campus, includes specialized training, joint lectures, and international symposia. Students gain essential skills for independent infection research, with hands-on courses in handling and working with BSL2-4 pathogens. The program emphasizes inclusivity, interdisciplinary exchange, and international networking, particularly with African partner sites.
PhD positions
We are looking forward to applications from individuals with:
- An excellent master’s degree or equivalent in biology, physics, biochemistry, chemistry, molecular life sciences, infection biology or a related subject
- A high motivation and a strong interest in the combination of structural biology, immunology and virology
- Conscientiousness, a structured working approach and a strong work ethic
- Commitment, team spirit and good communication skills, independence and flexibility
- Very good spoken and written English
We offer several PhD positions with a salary of 65% grade 13 (TV-L E13) and a duration of three and a half years with social security and health benefits.
We provide a diverse and equal opportunity work environment regardless of age, gender, sexual identity, disability, race or religion. Our institutions have signed and commit to the Charta of Diversity. We explicitly aim to increase the proportion of women in management positions, especially among academic staff in research and teaching. Women with equal qualifications will be given preference. The same applies in the case of under-representation of one gender in the area of employment. Persons with severe disabilities with equal aptitude, competence and professional performance will be given priority.
Does that sound interesting? Then get in touch with us.
Send an email to crc1648@uke.de including (as a single PDF file):
- Letter of motivation indicating you area of interest and/or preferred project
- CV including two references - Certificates (University entrance, qualification, BSc, and MSc)
- Abstract of Master thesis (1 page) including a summary of experimental skills
- List of publications (if applicable)
We look forward to receiving your email until July 14th, 2024.
Interested candidates are welcome to express interest in several projects/institutes within CRC 1648. Furthermore, we would like to distribute applications within the CRC 1648 consortium. If this is not desired, please indicate this in your mail.
PhD Projects
-
Project A02: Humanized mice to study the zoonotic potential and pathophysiology of hepatotropic emerging pathogens
Project A02: Humanized mice to study the zoonotic potential and pathophysiology of hepatotropic emerging pathogens
A number of emerging viruses have been associated with liver disease, including new hepatitis B-like viruses from bats, and other not strictly hepatotropic viruses such as EBOV, DENV and YFV. We hypothesize that the capacity of emerging viruses to replicate in liver cells impacts pathophysiology and disease outcome. We will develop and use distinct humanized and chimeric mouse models to understand zoonotic potential, hepatotropism and pathophysiology of these viruses, and establish models to study yet unknown emerging pathogens (Pathogen ‘X’).
Project leaders:
Prof. Dr. Maura Dandri, UKE
Dr. Estefanía Rodríguez, BNITM -
Project A05: Pathophysiology of severe manifestations of Lassa fever
Project A05: Pathophysiology of severe manifestations of Lassa fever
Lassa fever (LF) is recognised by the WHO as a priority disease for urgent R&D. About 20% of LF patients die in hospitals in West Africa. We hypothesise that severe Lassa fever presents either as septic shock syndrome with acute kidney injury, hyperinflammation, impaired coagulation and endothelial dysfunction or as an acute neurological condition with seizures and encephalopathy. We will conduct comprehensive clinical, biomarker, human tissue and organoid studies to elucidate these pathophysiological features with the aim of developing host-specific therapies.
Project leaders:
Dr. med. Till Omansen, UKE, BNITM
Prof. Dr. Michael Ramharter, UKE, BNITM
Prof. Dr. Stephan Günther, BNITM -
Project B03: Structural cell biology of Bunyaviruses
Project B03: Structural cell biology of Bunyaviruses
Bunyaviruses cause regular outbreaks and therefore pose an imminent threat to global public health. Our understanding of the biology of these viruses is however limited. Although structures of the isolated bunyavirus L protein, a key player in viral genome replication and transcription processes, have been solved, knowledge on the broader context of these processes is needed to understand the involved host and viral factors and their spatiotemporal functional orchestration inside cells. By obtaining information on protein-protein interfaces we will be able to identify novel Achilles heels for the development of antiviral strategies.
Project leaders:
Dr. Maria Rosenthal, CSSB, BNITM
Prof. Dr. Kay Grünewald, CSSB, LIV, UHH -
Project B04: Determinants of heterospecific phase separation in negative-strand RNA viruses
Project B04: Determinants of heterospecific phase separation in negative-strand RNA viruses
The goal of B04 is to create broad-spectrum antivirals against negative-strand RNA viruses like Ebola and Lassa and a potential disease X by inhibiting viral liquid-liquid phase separation, resulting in viral inclusion body formation, which is crucial for replication. Using a novel live-cell Click-approach for viral RNA labeling, we will identify viral and cellular proteins in functional viral condensates. We will employ neural networks and wet lab confirmation to decode the sequence “grammar” driving this recruitment. This approach will guide the development of inhibitors targeting viral condensate formation as a new class of antivirals.
Project leaders:
Prof. Dr. Jens Bosse, CSSB, MHH
Prof. Dr. Chris Meier, UHH, CSSB -
Project C01: Protective early response patterns of innate cellular immune crosstalk in hemorrhagic fever virus infection
Project C01: Protective early response patterns of innate cellular immune crosstalk in hemorrhagic fever virus infection
The main hypothesis is that strong activation of natural killer (NK) cells prevents viral spread and excessive tissue inflammation in viral haemorrhagic fevers. Using closely related pairs of pathogenic and apathogenic arena- and filoviruses, we will investigate how infected macrophages activate NK cells and what factors render them resistant to NK cell killing. We will use cerebral organoids to study the brain-specific macrophage response to viral infection, as rapid control of inflammation is essential in this tissue. The long-term goal is to identify targets for NK cell-based therapies to improve viral control and resolve inflammation.
Project leader:
-
Project C04: Multidimensional analysis of protective and damaging adaptive immune responses in emerging viral infections
Project C04: Multidimensional analysis of protective and damaging adaptive immune responses in emerging viral infections
The ability to quickly analyze host immune response patterns to novel viral infections and mechanisms of immune evasion are key for pandemic preparedness. Thus, we focus on analyzing host immune responses to emerging viruses. We hypothesize that initial naïve adaptive immune responses, while aiding in pathogen clearance, might lead to hyperinflammation or autoimmune-like reactions. Additionally, we aim to examine the switch from innate to adaptive immunity, especially in immunocompromised and re-infected vaccinated patients to gain insights into immune dynamics.
Project leaders:
Prof. Dr. Mascha Binder, Universitätsspital Basel, UKE
Prof. Dr. Julian Schulze zur Wiesch, UKE
-
Project D01: Characterization of host factors as targets for the development of broadly acting therapeutics against emerging viruses
Project D01: Characterization of host factors as targets for the development of broadly acting therapeutics against emerging viruses
This project addresses two parts to design broad-spectrum active molecules. Highly active and selective inhibitors against the host-cell enzyme DHODH will be developed. DHODH is essential for the de-novo-biosynthesis of pyrimidine nucleosides. Moreover, nucleoside analogue triphosphates will be synthesized and evaluated against viral RNA-polymerases. Inhibitory triphosphates will be converted into their pronucleotide forms to enable membrane-permeability. The project aim is to block two essential enzymes as a new combinational intervention approach also applicable to tackle “Disease X” in the future.
Project leader:
-
Project D02: Metagenomics in emerging viral infections – integrating non-host and host transcriptional responses
Project D02: Metagenomics in emerging viral infections – integrating non-host and host transcriptional responses
D02 seeks to advance pathogen detection through metagenomic and metatranscriptomic methods, refining next-generation sequencing approaches. It involves cataloging microbial and host-derived sequences, creating expression signatures for specific diagnostic entities, and developing host signatures for viral families. Leveraging expertise in viral genomics, the project integrates nucleotide sequence-independent methods for pathogen detection. Furthermore, it aims to predict infectious diseases by analyzing disease-associated host responses, addressing challenges posed by unknown pathogens like “Disease X”.
Project leader:
-
Project D03: Dissecting protective immunity to viral vector vaccines for emerging viruses
Project D03: Dissecting protective immunity to viral vector vaccines for emerging viruses
Vaccines represent one of the world’s most impactful public health interventions, resulting in the prevention of millions of infections and deaths annually. Viral vector vaccines (VVV) have been developed in response to recent emerging virus outbreaks. In this project we investigate immunity to two VVV, MVA-MERS and VSV-EBOV, using human biomaterial from investigator initiated clinical trials. We specifically analyse frequently understudied immune parameters, including non-neutralizing antibody responses, T cell immunity and the impact of vector immunity on VVV-induced immune response.
Project leader:
MD positions
The CRC 1648 offers several scholarships per year for an experimental medical doctoral thesis at the participating institutions.
We are looking forward to applications from individuals with:
- A high motivation and a strong interest in the combination of structural biology, immunology and virology
- Conscientiousness, a structured working approach and a strong work ethic
- Commitment, team spirit and good communication skills, independence and flexibility
- Very good spoken and written English
- Laboratory experience would be desirable
Start: Flexible by arrangement. The work requires two semesters off/one research year. A scholarship will be paid for this period.
We look forward to receiving your application with CV and short letter of motivation to
crc1648@uke.de